Overview
A reproducible, treatment-aware survival model for identifying robust genomic predictors in breast cancer.
This case study summarizes a full survival analysis of 1,047 women from the TCGA-BRCA cohort. We combine RNA-Seq expression with clinical covariates and treatment indicators to uncover a small, stable subset of genes that truly matter for overall survival.
High-dimensional modeling is carried out using the Cox proportional hazards framework, treatment–gene interactions, and a 2,000-seed LASSO stability-selection pipeline designed to filter out unstable features and retain only reproducible signals.
Methods in Brief
Survival time \(t\) is modeled using a Cox proportional hazards model with clinical, treatment, and gene-expression covariates:
$$ h(t \mid x) = h_0(t)\exp(\beta^\top x). $$
The workflow:
- Preprocess clinical data and normalize RNA-Seq expression.
- Univariate Cox screening with effect-size filtering.
- LASSO-Cox modeling across 2,000 deterministic seeds to quantify the selection stability of each gene.
- Final multivariable Cox model integrating stable genes and treatment–gene interactions.
Stability Across 2,000 LASSO Models
Most genes appear in only a handful of LASSO models, indicating low stability. A much smaller subset appears consistently across hundreds of seeds—these form the stable core carried into the final model.
This stability-selection process dramatically reduces false positives and yields reproducible biomarkers.

Final Model: Stable Genes and Treatment Effects
The final Cox model includes clinical covariates, treatment indicators, and the most stable genes identified by the LASSO process. Hazard ratios greater than 1 increase risk; values below 1 indicate protective effects.
Treatment–gene interactions reveal that certain genes amplify or diminish the benefit of therapy, underscoring the importance of a treatment-aware modeling strategy.
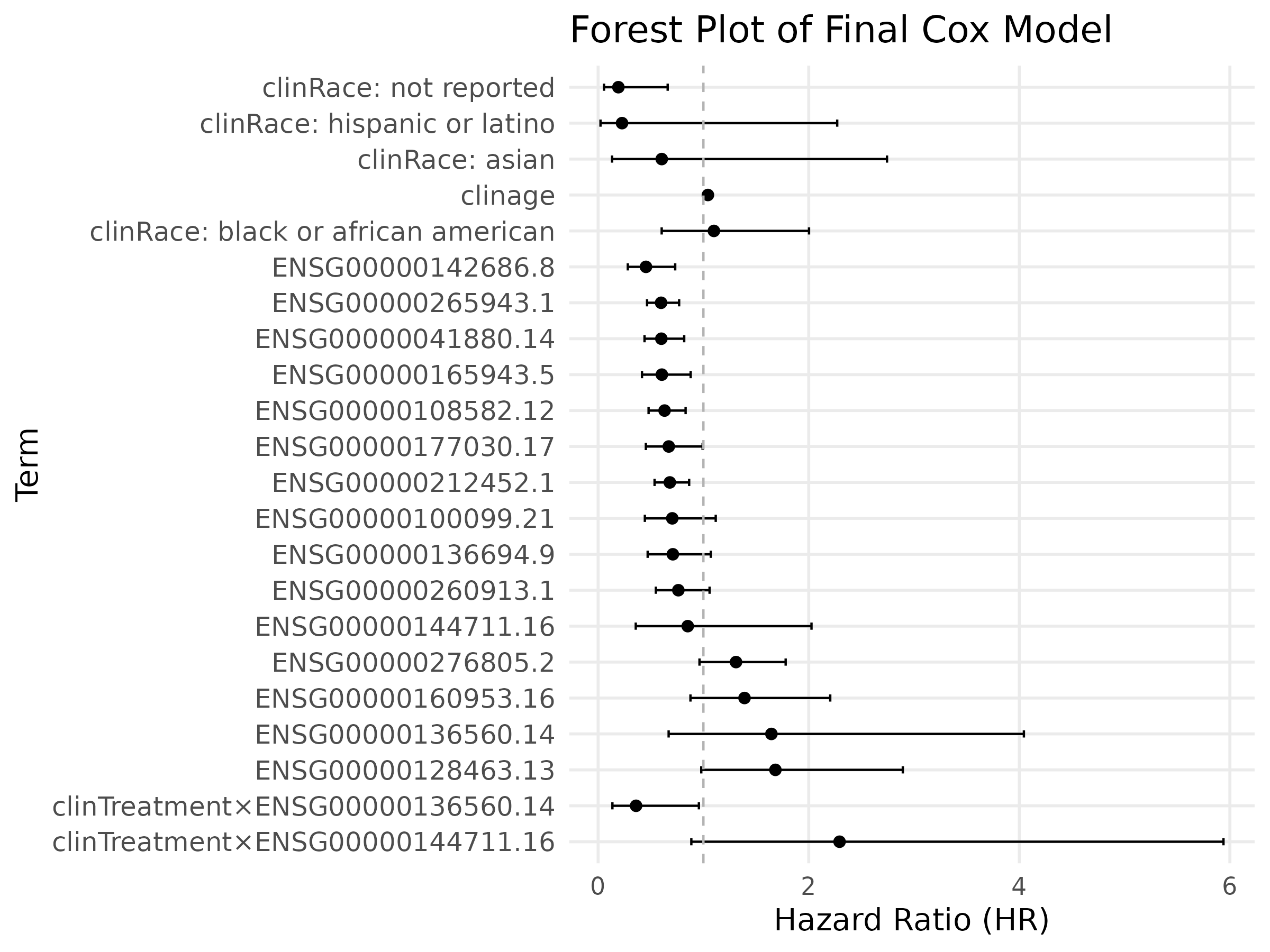
Selected Genes That Matter
Three representative stable genes are shown below. High-expression groups (red) and low-expression groups (blue) exhibit distinct survival trajectories.
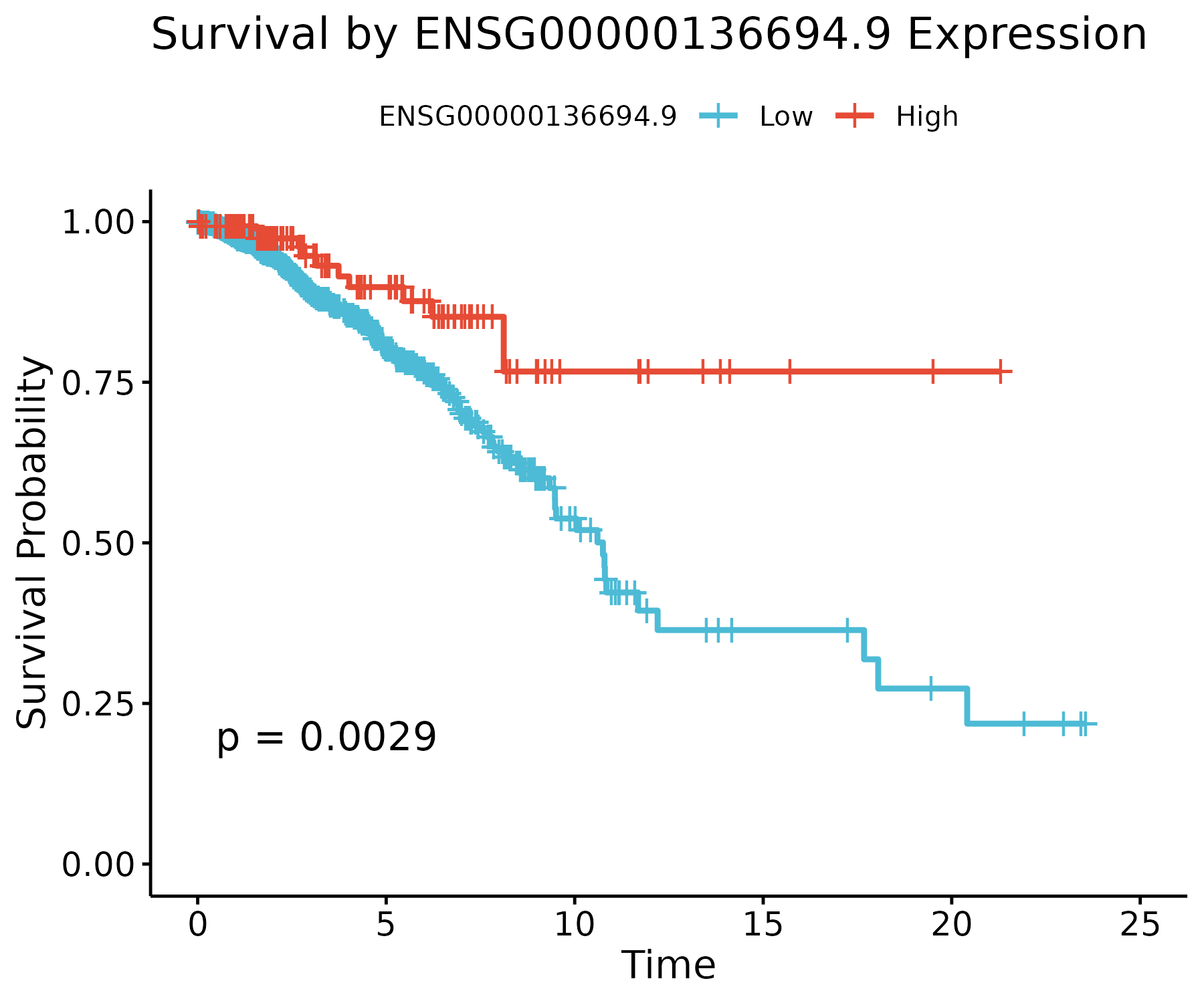
ENSG00000136694.9
Shows one of the strongest separations in the dataset: high expression corresponds to substantially improved survival (p ≈ 0.003).
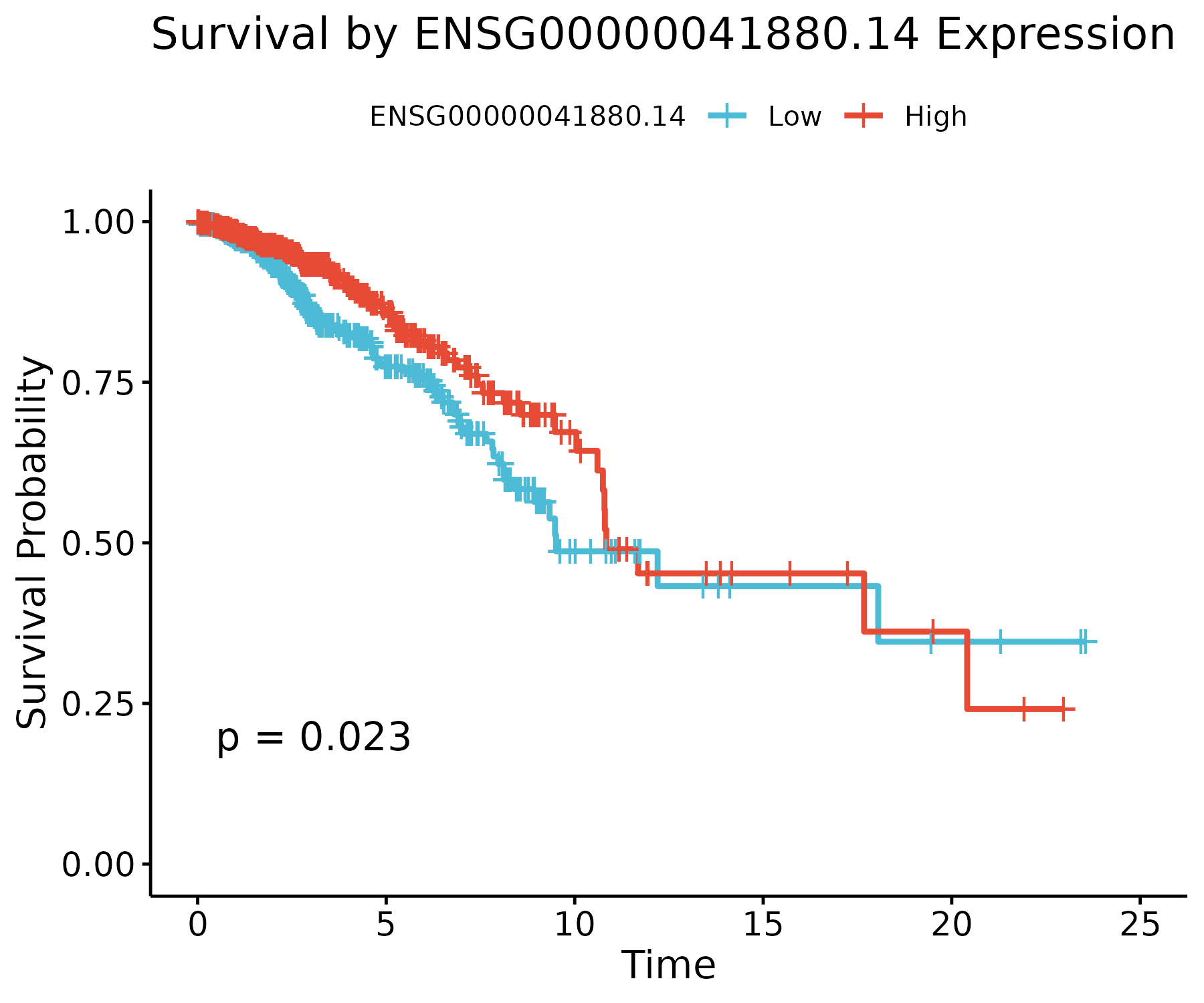
PARP3 (ENSG00000041880.14)
A DNA-damage–response gene within the PARP family. Higher expression is linked to improved survival (p ≈ 0.02), consistent with known DNA repair pathways.
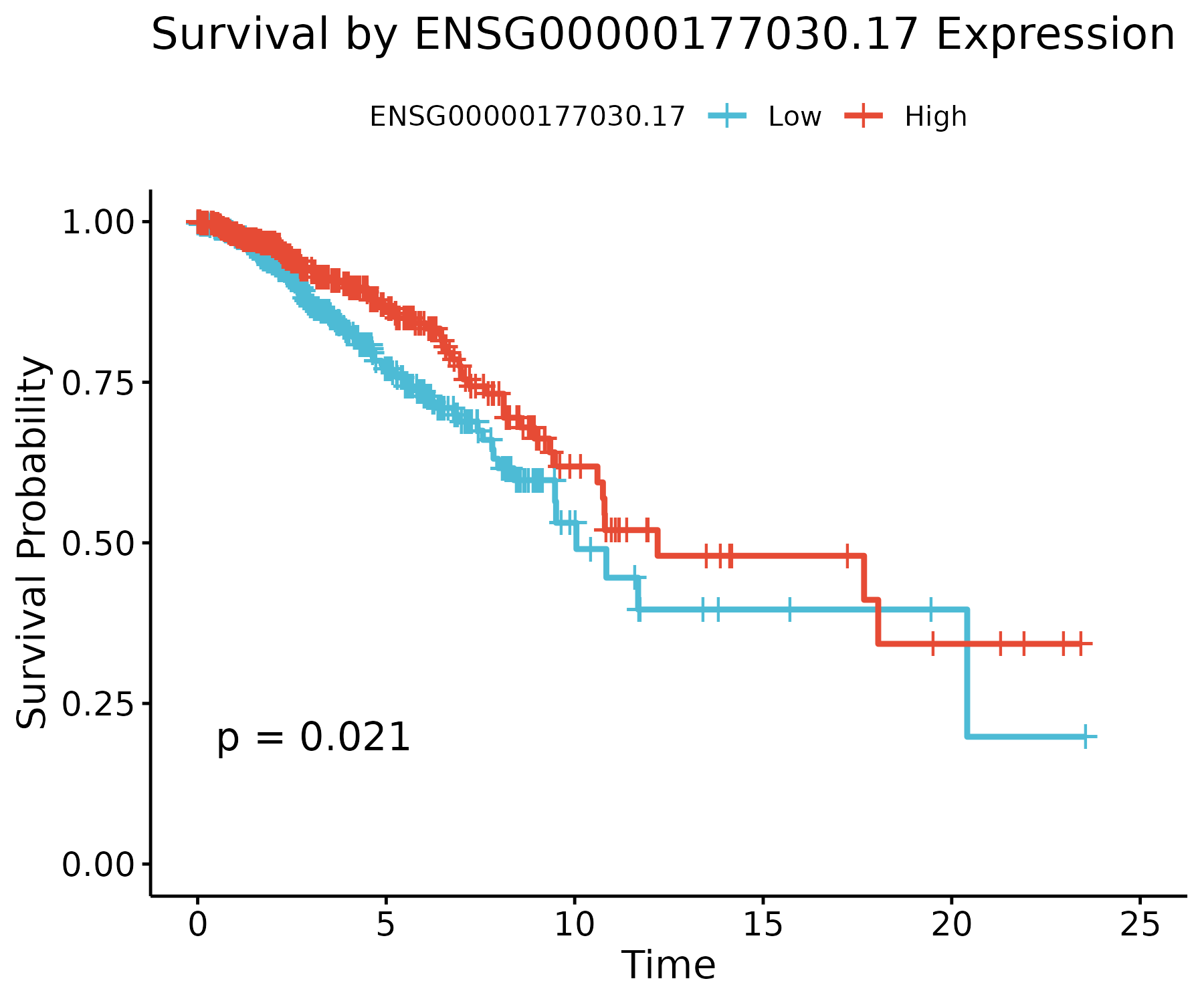
DEAF1 (ENSG00000177030.17)
An immune-regulatory transcription factor. Its expression stratifies survival significantly (p ≈ 0.02), highlighting the role of immune signaling.
Takeaways
Stability-selection and treatment-aware modeling reduce tens of thousands of genes to a concise, reproducible set with strong prognostic value. The framework generalizes naturally to other cancers and datasets.
Resources
Full methodological details, diagnostics, and extended figures are available in the complete manuscript.
Download full manuscript (PDF)
Contact
Interested in applying similar pipelines to your data? contact@midnightmechanism.xyz